metaGOflow: A workflow for marine Genomic Observatories data analysis
How to answer fast the question "who is out there"?
What is this project about?
Genomic Observatories (GOs) are sites subjected to long-term scientific research, including the study of genomic biodiversity, from single-celled microbes to multicellular organisms. GOs, under the European Marine Omics Biodiversity Observation Network (EMO BON), a European Marine Biological Resource Centre (EMBRC) initiative, will be providing regular monthly (or bimonthly) sampling along with shotgun metagenomics data of microbial communities from marine sites throughout Europe. However, the success of GOs, and of similar effots, lies in the prompt supply of the marine biological community with FAIR (Findable, Accessible, Interoperable, and Reusable) data but also with preliminary, robust and standardized, results from their analysis.
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 824087, EOSC-Life, and more specifically, under the Digital Life Sciences Open Call.
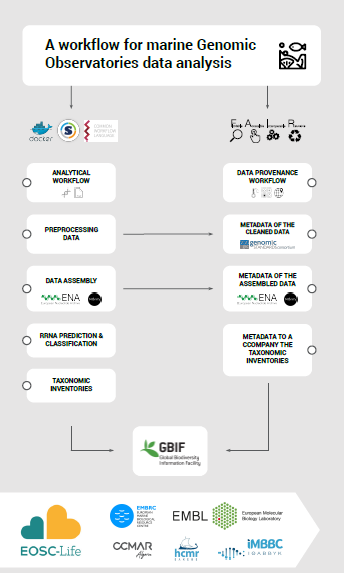
The overall focus of the project is to build an effective workflow for the analysis of the metagenomic GO data in a time-effective and reproducible way. Based on the well established MGnify resource, we developed a workflow to address these challenges combining the Common Workflow Language (CWL) notion with the Research Object Crate (RO-Crate)concept.
metaGOflow supports the fast inference of the taxonomic profiles of the GO-derived data based on rRNA genes and their functional annotation using its raw reads.
metaGOflow was developed as a joint project by ELIXIR (EMBL-EBI) and seven national nodes of the European Marine Biological Resource Centre (EMBRC) (BE, UK, FR, GR, IT, PT, ES) with the ultimate mission to build a distributed workflow for analyses of marine metagenomic data generated by EMBRC’s GOs.
More information: metaGOflow